from pathlib import Path
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as snsData visualization
Insurance Claim Analysis
The task at hand is to identify health and demographic characteristics that lead to poor health, using health insurance claim amounts as an indicator.
Data sources: - Kaggle - data.world
Config
# columns in the data
TARGET_COL = "claim"
# plots
%matplotlib inline
sns.set_theme(context="notebook", style="whitegrid", rc={"figure.figsize": (14, 8)})Loading the data
# Google Drive link: https://drive.google.com/file/d/18zxQ8rwoinnWBTcDxhgP7pO_3QUSWcpZ/view?usp=sharing
df = pd.read_csv(
"https://drive.google.com/uc?id=18zxQ8rwoinnWBTcDxhgP7pO_3QUSWcpZ",
index_col="index"
)
df.info()<class 'pandas.core.frame.DataFrame'>
Int64Index: 1340 entries, 0 to 1339
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PatientID 1340 non-null int64
1 age 1335 non-null float64
2 gender 1340 non-null object
3 bmi 1340 non-null float64
4 bloodpressure 1340 non-null int64
5 diabetic 1340 non-null object
6 children 1340 non-null int64
7 smoker 1340 non-null object
8 region 1337 non-null object
9 claim 1340 non-null float64
dtypes: float64(3), int64(3), object(4)
memory usage: 115.2+ KBYData profiling report
import sys
!{sys.executable} -m pip install -q ydata-profilingfrom ydata_profiling import ProfileReport
report = ProfileReport(df)
# uncomment the line below to see the report
# reportSeparate features from target
y = df[TARGET_COL]
X = df.drop(TARGET_COL, axis=1)
numeric_dtypes = ["int64", "float64"]
categorical_df = X.select_dtypes(exclude=numeric_dtypes)
numeric_df = X.select_dtypes(include=numeric_dtypes)Distribution of variables
Target variable
fig, ax = plt.subplots()
sns.histplot(x=y, ax=ax, log_scale=True, kde=True)
ax.set_title(f"Distribution of {TARGET_COL}")
plt.show()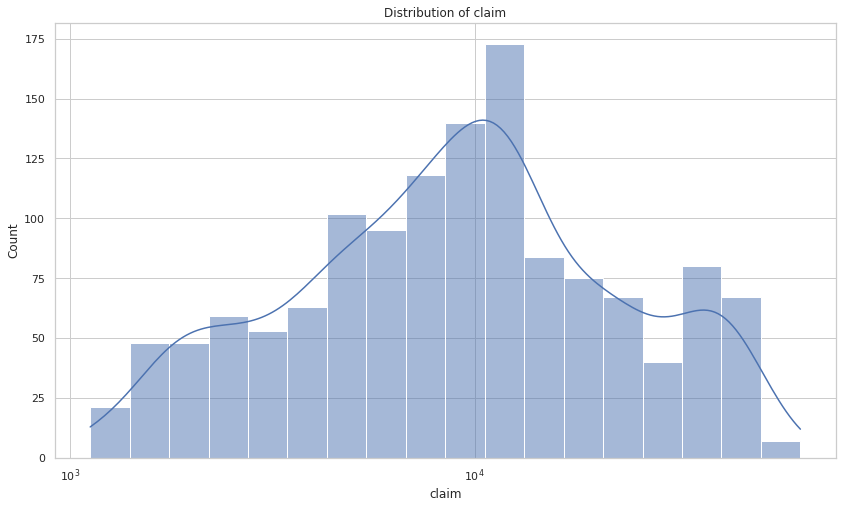
Numeric features
cols = 2
rows = np.ceil(numeric_df.shape[1] / cols).astype(int)
fig, axes = plt.subplots(rows, 2, figsize=(14, 8 // cols * rows))
plt.tight_layout()
for i, col in enumerate(numeric_df.columns):
ax = axes[i // cols, i % cols]
sns.histplot(data=df, x=col, ax=ax)
ax.set_title(f"Histogram of {col}", y=0.88)
plt.show()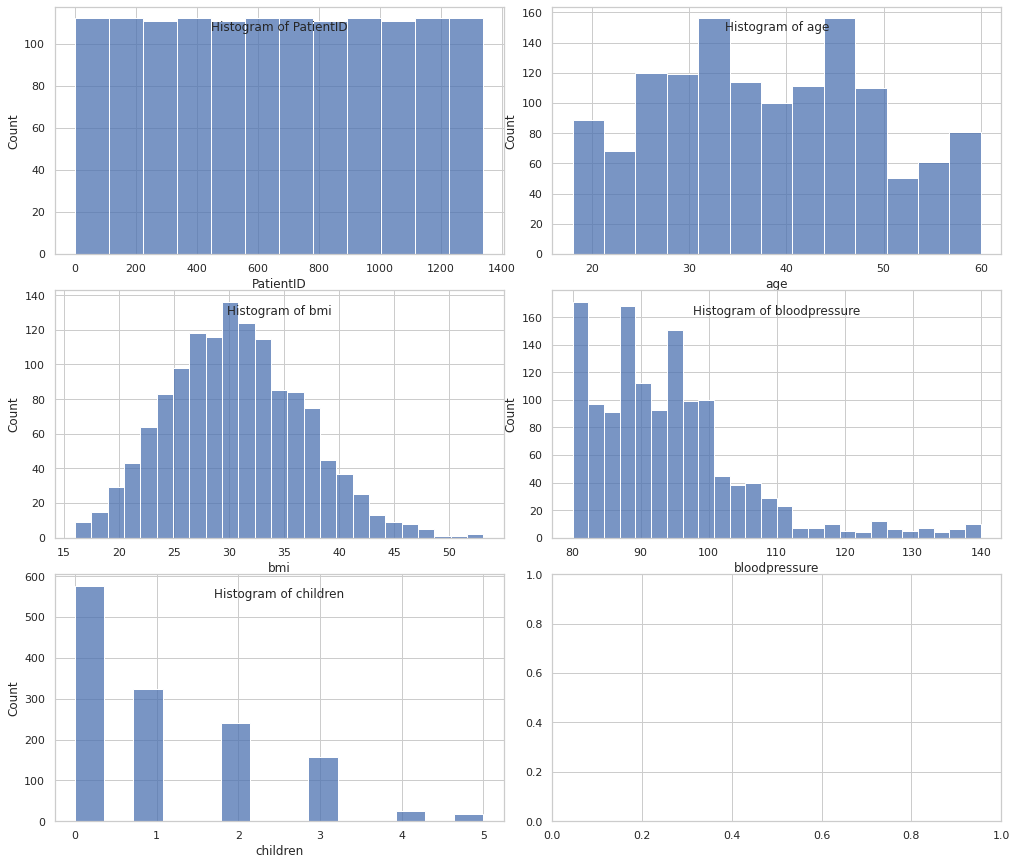
Numeric features by target
cols = 2
rows = np.ceil(numeric_df.shape[1] / cols).astype(int)
fig, axes = plt.subplots(rows, 2, figsize=(14, 8 // cols * rows))
plt.tight_layout()
for i, col in enumerate(numeric_df.columns):
ax = axes[i // 2, i % 2]
sns.scatterplot(data=df, x=col, y=TARGET_COL, ax=ax)
ax.set_title(f"Scatter plot of {col} by {TARGET_COL}", y=0.88)
plt.show()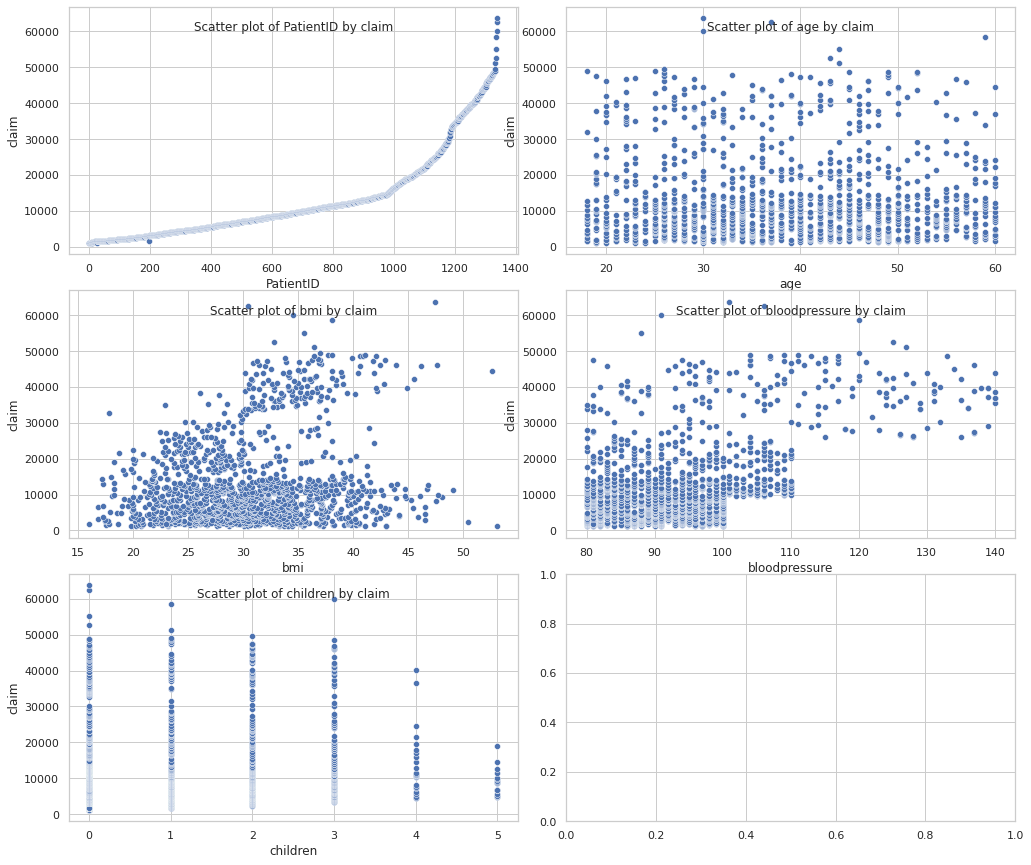
Categorical features
for col in categorical_df.columns:
display(X[col].value_counts(normalize=True).to_frame())| gender | |
|---|---|
| male | 0.50597 |
| female | 0.49403 |
| diabetic | |
|---|---|
| No | 0.520896 |
| Yes | 0.479104 |
| smoker | |
|---|---|
| No | 0.795522 |
| Yes | 0.204478 |
| region | |
|---|---|
| southeast | 0.331339 |
| northwest | 0.261032 |
| southwest | 0.234854 |
| northeast | 0.172775 |
cols = 2
rows = np.ceil(categorical_df.shape[1] / cols).astype(int)
fig, axes = plt.subplots(rows, 2, figsize=(14, 8 // cols * rows))
plt.tight_layout()
for i, col in enumerate(categorical_df.columns):
ax = axes[i // 2, i % 2]
sns.countplot(data=X, x=col, ax=ax)
ax.set_title(f"Count of {col} classes", y=0.88)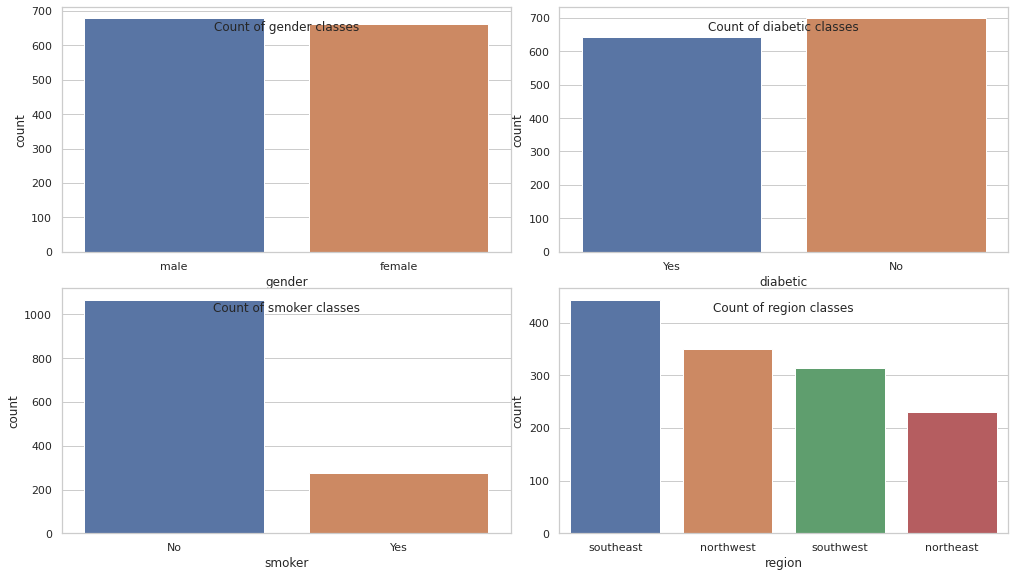
Categorical features by target
cols = 2
rows = np.ceil(categorical_df.shape[1] / cols).astype(int)
fig, axes = plt.subplots(rows, 2, figsize=(14, 8 // cols * rows))
plt.tight_layout()
for i, col in enumerate(categorical_df.columns):
ax = axes[i // 2, i % 2]
sns.stripplot(data=df, x=col, y=TARGET_COL, hue=col, ax=ax)
ax.set_title(f"Scatter plot of {TARGET_COL} by {col}", y=0.88)
plt.show()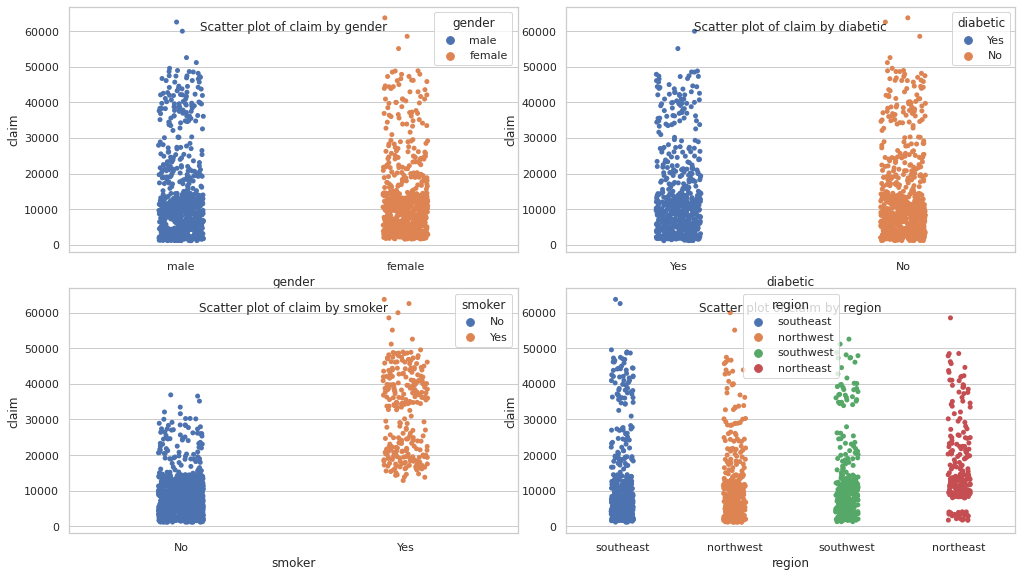
cols = 2
rows = np.ceil(categorical_df.shape[1] / cols).astype(int)
fig, axes = plt.subplots(rows, 2, figsize=(14, 8 // cols * rows))
plt.tight_layout()
for i, col in enumerate(categorical_df.columns):
ax = axes[i // 2, i % 2]
sns.boxplot(data=df, x=col, y=TARGET_COL, ax=ax)
ax.set_title(f"Box plot of {TARGET_COL} by {col}", y=0.88)
plt.show()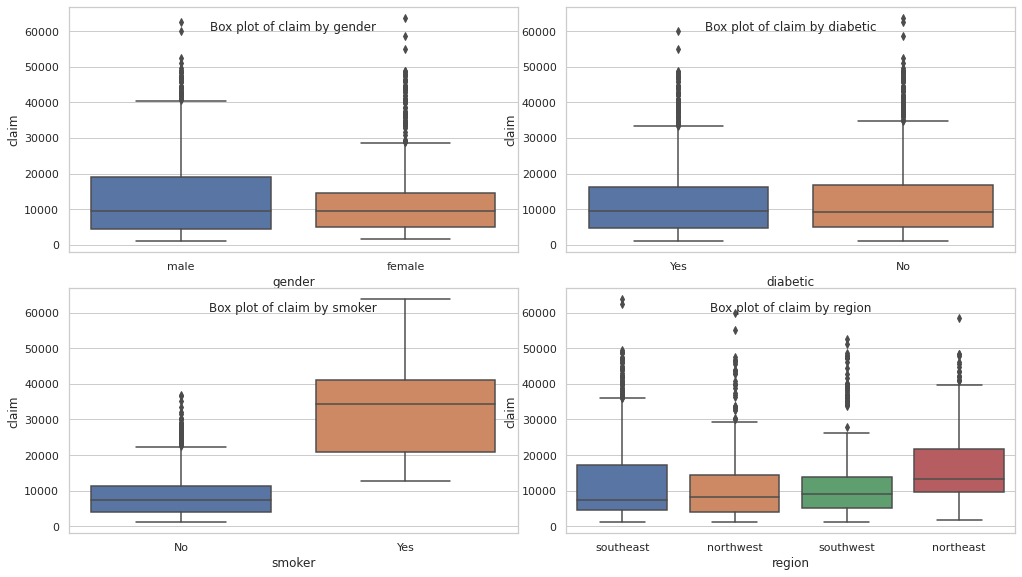
TODO
- Discretize blood pressure. Refer to https://www.heart.org/en/health-topics/high-blood-pressure/understanding-blood-pressure-readings
- Discretize BMI. Reference: https://www.cdc.gov/healthyweight/assessing/bmi/adult_bmi/index.html#InterpretedAdults